CSV File
In MEDimage every dataset must have a csv file along with it, this file contains information
about the scans in the dataset that will be used in the radiomics analysis, especially region of interest (ROI) names used in
each scan. Since scans can have multiple regions of interest (ROIs), the user needs to specify for each scan the ROI name(s) to use
for the processing and radiomics extraction. The csv files are also used by the DataManager in pre-checks and
summary creation (after raw data processing). The different columns of the file are:
PatientID: The scan patient ID, for example:
"Glioma-TCGA-001".ImagingScanName: Type of the imaging modality, usually
"CT"for CT scans,"PT"for PET scans and MRI sequence for MR scans ("T1","T2"…).ImagingModality: Imaging modality (
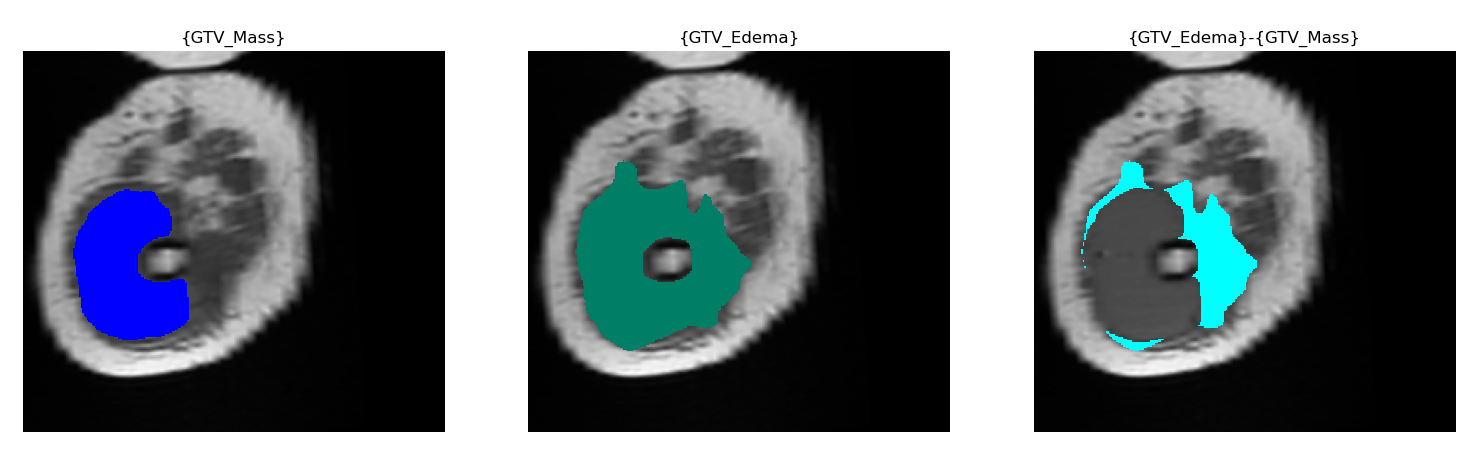
"CTscan","MRscan"or"PTscan").ROIname: ROI name for the analysis. Either addition or subtraction of multiple ROI names or a single ROI name. Every ROI name is put between brackets then added or subtracted to other ROI names. For example:
"{GTV_Edema}-{GTV_Mass}", which means"GTV_Mass"will be subtracted from"GTV_Edema"in the analysis. The following picture shows the result of this subtraction:
Note
The ROIname must be the same for all the scans in the csv file because scientifically, we process the same
ROI in a radiomics analysis.
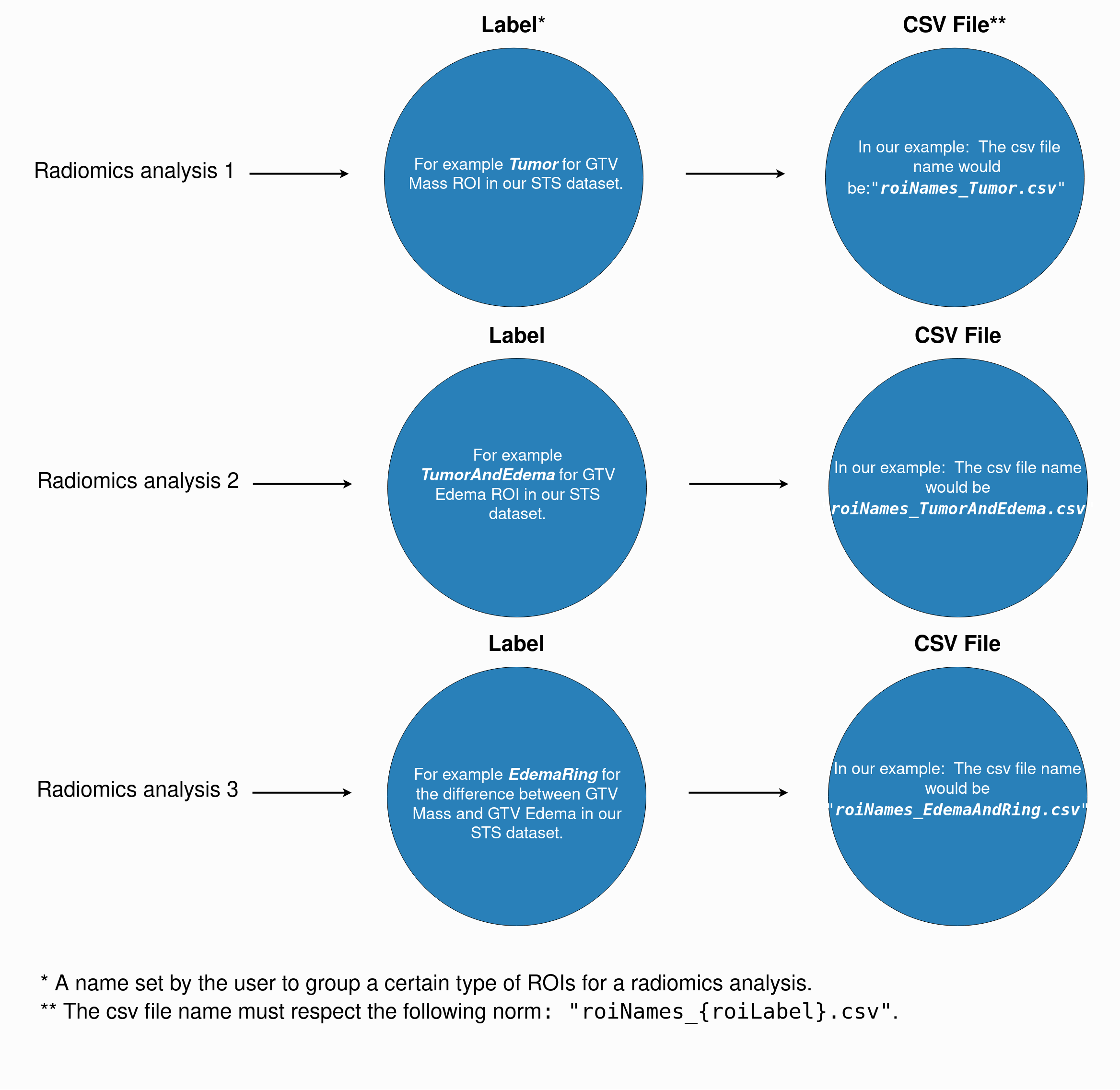
The csv files must respect the following naming norm: "roiNames_{roiLabel}.csv". The following figure gives a detailed example on how to choose
your dataset ROI label and how to name your csv files:
The following tables are an example of csv files for the same dataset Soft-Tissue-Sarcoma (STS) cancer consisting of different modalities (MR, CT and PET), different ROIs (GTV mass and GTV edema) and each table is for different radiomcis analysis:
Radiomics analysis 1:
"Tumor".
PatientID |
ImagingScanName |
ImagingModality |
ROIname |
|---|---|---|---|
STS-McGill-001 |
T1 |
MRscan |
{GTV_Mass} |
STS-McGill-001 |
T2 |
MRscan |
{GTV_Mass} |
STS-McGill-002 |
CT |
CTscan |
{GTV_Mass} |
STS-McGill-003 |
PET |
PTscan |
{GTV_Mass} |
Radiomics analysis 2:
"TumorAndEdema".
PatientID |
ImagingScanName |
ImagingModality |
ROIname |
|---|---|---|---|
STS-McGill-001 |
T1 |
MRscan |
{GTV_Edema} |
STS-McGill-001 |
T2 |
MRscan |
{GTV_Edema} |
STS-McGill-002 |
CT |
CTscan |
{GTV_Edema} |
STS-McGill-003 |
PET |
PTscan |
{GTV_Edema} |
Radiomics analysis 3:
"EdemaRing".
PatientID |
ImagingScanName |
ImagingModality |
ROIname |
|---|---|---|---|
STS-McGill-001 |
T1 |
MRscan |
{GTV_Edema}-{GTV_Mass} |
STS-McGill-001 |
T2 |
MRscan |
{GTV_Edema}-{GTV_Mass} |
STS-McGill-002 |
CT |
CTscan |
{GTV_Edema}-{GTV_Mass} |
STS-McGill-003 |
PET |
PTscan |
{GTV_Edema}-{GTV_Mass} |
Note
It is pointless in our case but it’s possible to analyze the addition of multiple ROIs, for example: "{GTV_Edema}+{GTV_Mass}".
Future works of MEDimage will aim to automate the creation of these csv files for each dataset and to implement ROIs intersection as well.