Input Data
MEDimage package accepts two formats of input data: NIfTI
and DICOM. Each format has its own conventions
that need to be followed. The following sections describe the norms and the conventions for each format and we recommend you process your
dataset in a way that respects them.
DICOM
Image
Every DICOM file contains a header and a body. The header contains the metadata of the image, and the body contains the image itself. The header contains information about the scan and the most important for our package is the following:
Patient ID: Primary identifier for the Patient, referenced in the
(0010,0020) PatientIDfield of the header. This field should not contain any underscore and for compatibility with other MEDomics packages, we recommend using the following format:'study-institution-numericID'. For example,'STS-McGill-001'. It is also used in the CSV File of the dataset under the columnPatientID.Series description: Referenced in the
(0008,103E) Series Descriptionfield of the DICOM header. A description of the series, usually describes the type of the modality used. This field must be renamed to be the same for each sequence of each modality. For example,'T1'for all the T1-weighted MRI scans and'T2'for all the T2-weighted MRI scans. It is referred to in the CSV File of the dataset asImagingScanName.
RTstruct
RTstruct files define the area of significance and hold information about each region of interest (ROI). The RTstruct files are associated with their imaging volume using the
(0020,000E) Series Instance UIDor the(0020,0052) Frame of Reference UIDfound in the file’s header.MEDimagepackage recommends the following:
Patient ID: Same conventions and recommendations as the DICOM image.
Series description: Same conventions and recommendations as the DICOM image.
ROI name: Only found in DICOM RTstruct files and referenced in each element (each ROI) of the
(3006,0020) Structure Set ROI Sequencelist of the DICOM header, under the attribute(3006,0026) ROI Namewhich is a name given to each region of interest (ROI).MEDimagehas no conventions over this field, but we recommend renaming each ROI name in a simple and logic way to differentiate them from each other. It is very important to keep track of all the ROIs in your dataset since they need to be specified in the CSV File of the dataset under theROINamecolumn to be used later in your radiomics analysis.
NIfTI
The NIfTI format is a simple format that only contains the image itself. Unlike DICOM, the NIfTI format does contain any
information about the regions of interest (ROI) so it needs to be provided in other separate files. In order for MEDimage to read a NIfTI scan
files, they need to be put in the same folder with the following names:
'PatientID__SeriesDescription(ROILabel).Modality.nii.gz': The image itself. For example:'STS-McGill-001__T1(GTV).MRscan.nii.gz'.'PatientID__SeriesDescription(ROIname).ROI.nii.gz': The ROI or the mask of the image. This file should contain a binary mask of the ROI. For example:'STS-McGill-001__T1(GTV_Mass).ROI.nii.gz'.
The following figure sums up the MEDimage logic in reading data for both formats:
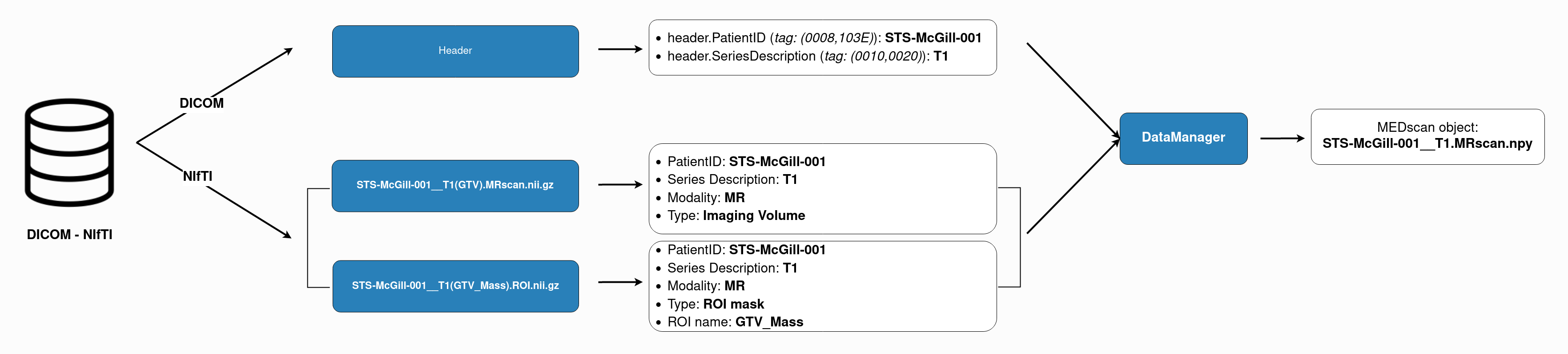
If these conventions are followed, the DataManager class will be able to read the data and create the MEDscan objects that will be used
in the radiomics analysis with no further intervention from the user. For instance, MEDimage package is capable of automatically updating
the fields of all the DICOM files as long as the dataset is organized in the following way:
dataset_folder
├── Patient ID 1
│ ├── ImagingScanName 1
│ │ ├── DICOM files
│ │ └── ...
│ └── ImagingScanName 2
│ ├── DICOM files
│ └── ...
├── Patient ID 2
│ ├── ImagingScanName 1
│ │ ├── DICOM files
│ │ └── ...
│ └── ImagingScanName 2
│ ├── DICOM files
│ └── ...
└── ...
For example:
dataset_folder
├── STS-McGill-001
│ ├── T1
│ │ ├── *.dcm
│ │ └── ...
│ └── PET
│ ├── *.dcm
│ └── ...
├── STS-McGill-002
│ ├── T2FS
│ │ ├── *.dcm
│ │ └── ...
│ └── CT
│ ├── *.dcm
│ └── ...
└── ...
Just run the following command:
python scripts/process_dataset.py --path-dataset path/to/your/dataset/folder
Note
Future works will include the automatic pre-processing of datasets according to the package conventions.